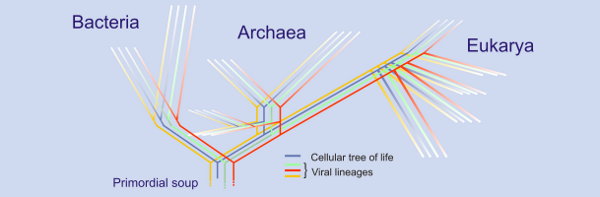
 |
| Putative tree of life including viral sequences. The 'sea' of viruses. http://blogs.helsinki.fi/bamford-group/files/2008/06/vevo.png |
Physically and evolutionarily speaking we are bathed in a sea of viruses and virus-like elements. Over the last billion or so years these genetic parasites have affected the lives of all kinds of organisms worldwide with sometimes devastating consequences (and others somewhat more positive). An extremely conservative estimate of virus numbers is more likely in the billions and so a major issue is just exactly how can we make sense of all this diversity out there in the Virosphere?
Determining how one virus – or group of viruses – is related to any other is pretty straightforward nowadays; with sequencing technology and bioinformatic analysis commonplace, we can effortlessly compare nucleotide sequences and generate phylogenetic trees displaying how two or more relate to each other. This works pretty well in a modern setting when we can compare viral isolates from patients infected in a certain outbreak and determine the path of spread for example over days, weeks, months or years,
 |
| Phylogenetic tree tattoo. http://blogs.discovermagazine.com/loom/files/2008/07/origin.jpg |
This kind of analysis may however have its limits when we attempt to apply it to virus groups that have potentially diverged millions of years ago – a scenario which would be expected given that viruses have been around with us since the beginning of life itself. Simply over that length of time, any evolutionary ‘signal’ found within a particular gene sequence will have disappeared. As masters of genetic variation, virus genomes can get pretty messy, evolutionary speaking due to horizontal gene transfer, recombination and high mutation rates. How can we then assess the deeper evolutionary questions with regards these highly variable agents?
Krupovic and Bamford (who both specialise in the origins and early evolution of viruses), commenting in the Journal of Virology recently, suggest that we should be focussing our analysis at a different aspect of viral lifestyles: the structural biology of their virions – a feature which they believe reflects the true nature of what a virus really is. This, they say should be suggestive of the early and highly conserved evolutionary relationships (nucleotide sequence may be variable but protein sequence/structure will be more highly conserved). By adopting this strategy, the problems of horizontal gene transfer may also be ablated making all variation observed relative to this gene/protein architecture. Using this perspective, we could infer higher taxonomic structures for the Virosphere reflecting true, ancient biological relationships – that seemingly unrelated viruses infecting divergent host species share a common origin (which has been seen for some human and bacterial viruses).
| Picornavirus virion structure made up of viral capsid proteins. Illustration by David S. Goodsell of The Scripps Research Institute (see this site) [Public domain], via Wikimedia Commons |
By attempting to group distantly related viruses, maybe those infecting bacteria, archaea and eukaryotes into common taxonomic classes we should be able to bring more order and structure to the viral universe. – facilitating a better understanding of the entire tree of life. We will not, however be able to accomplish this unless we realise the true age of these viruses and adapt our evolutionary investigations for this. Caveats aside, the idea presented by Krupovic and Bamford should stimulate further evolutionary work to be carried out on more virus taxa.
Order to the Viral Universe. Krupovic and Bamford J. Virol..2010; 84: 12476-12479